random_forests
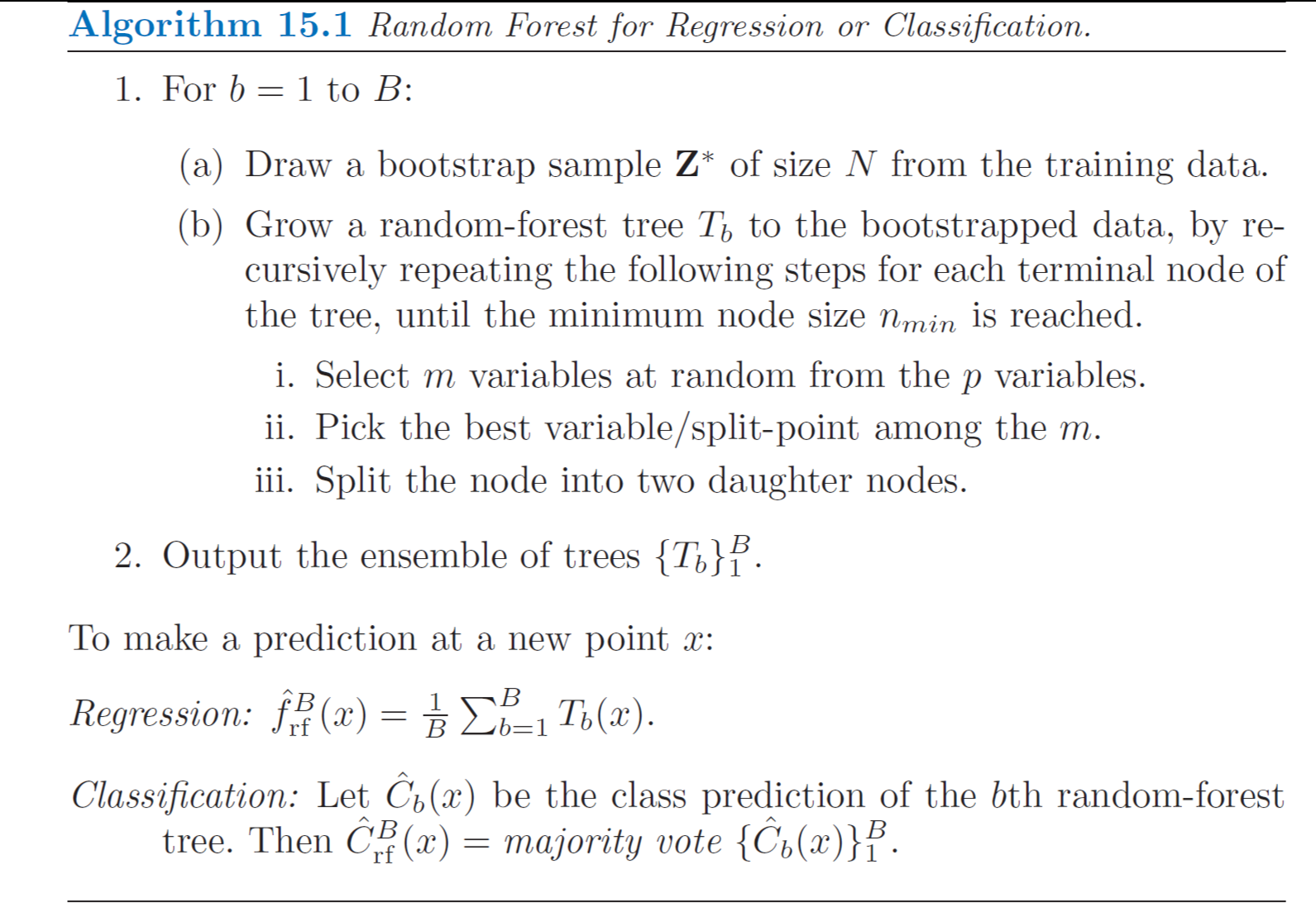
Random Forest
Bootstrap
Bagging
Earlier, we see that we can use bootstrap as a way of assessing the accuracy of a parameter estimate or a prediction. Here, we show how to use the bootstrap to improve the estimate or prediction itself.
Consider
Random Forest
Random Forests is a substantial modification of bagging that builds a large collection of de-correlated trees and then averages them. The essential idea in bagging is to average many noisy but approximately unbiased models and hence reduce the variance. Trees are ideal candidates for bagging, since they can capture complex interaction structures in the data, and if grown sufficiently deep, have relatively low bias. Since trees are notoriously noisy, they benefit greatly from the averaging (variance reduction).
Since each tree generated in bagging is identically distributed, the expectation of an average of \(B\) such tres is the same as the expectation of any one of them, this means the bias of bagged trees is the same as that of the individual trees and the only hope is the variance reduction. This is contrast to boosting, where the trees are grown in an adaptive way to remove bias, and hence are not identically distributed.
An average of \(B\) i.i.d random variables each with variance \(\sigma^2\), has variance \(\frac{1}{B} \sigma^2\). If the variables are simply i.d (not necessarily independent) with positive pairwise correlation \(\rho\), the variance of the average is:
\[\rho \sigma^2 + \frac{1 - \rho}{B} \sigma^2\]
As \(B\) increases, the second term disappears, but the first remains, and hence the size of the correlation of paris of bagged trees limits the benefits of averaging. The idea in random forest is to improve the variance reduction of bagging by reducing the correlation between the trees without increasing the variance too much. This is achieved in the tree-growing process through random selection of the input variables.
Specifically, when growing a tree on a bootstrapped dataset:
- Before each split, select \(m \leq p\) of the input variables at random as candidates for splitting (typically values for \(m\) are \(\sqrt{p}\) or even as low as 1)
- After \(B\) such trees \(\{T(\mathbf{x};\; \Theta_b)\}^{B}_{b=1}\) are grown, the random forest regression predictor is: \[\hat{f}^B_{rf} (\mathbf{x}) = \frac{1}{B}\sum^{B}_{b=1} T(\mathbf{x};\; \Theta_b)\]
- For classification, a random forest obtains a class vote from each tree, and then classifies using majority vote.
Out of Bag Samples
An importance feature of random forest is its use of out-of-bag samples:
- For each observation \(\mathbf{z}_i = (\mathbf{x}_i, y_i)\), construct its random forest predictor by averaging only those trees corresponding to bootstrap samples in which \(\mathbf{z}_i\) did not appear.
An out-of-bag error estimate (If short of data, it can be used as validation error) is almost identical to that obtained by \(N\)-fold cross validation, so unlike many other nonlinear estimators, random forest can be fit in one sequence, with cross-validation being performed along the way. Once the OOB error stabilizes, the training can be terminated.
Variable Importance
Random forests also use the OOB samples to construct a different variable-importance measure, apparently to measure the prediction strength of each variable. When the \(b\)th tree is grown, the OOB samples are passed down the tree, and the prediction accuracy is recorded. The values for the \(j\)th variable are randomly permuted in the OOB samples, and the accuracy is again computed. The decrease in accuracy as a result of this permuting is averaged over all trees, and is used as a measure of the importance of variable \(j\) in the random forest.
Overfitting
It is certainly true that increasing \(B\) does not cause the random forest sequence to overfit. like bagging, the random forest estimate approximates the expectation:
\[\hat{f}_{rf} (\mathbf{x}) = E[T(\mathbf{x}; \; \Theta)] = \lim_{B \rightarrow \infty} \hat{f}(\mathbf{x})^B_{rf}\]
with an average over \(B\) realizations of \(\Theta\). However, this limit can overfit the data, the average of fully grown trees can result in too rich a model and incur unnecessary variance. One possible solution is to reduce tree depth.