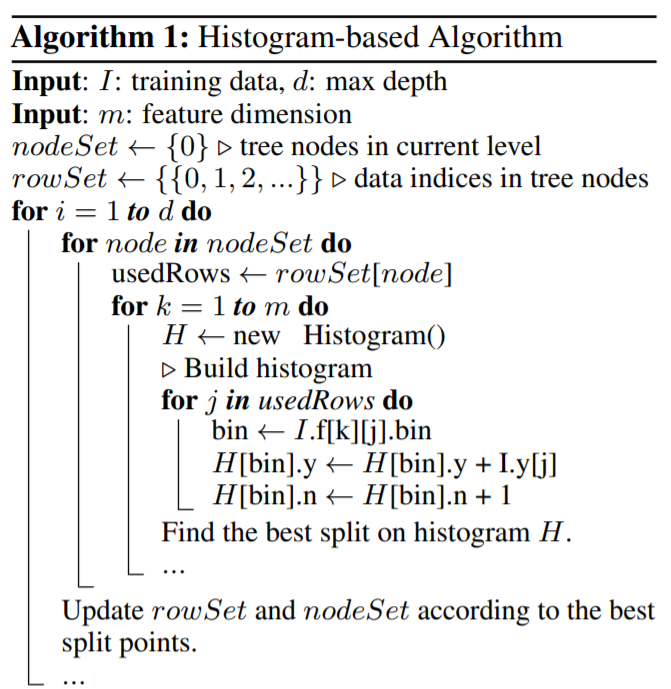
Graphical Models
Conditional Independence
Conditional Independence of Random Variable
Let \(\mathbf{X}, \mathbf{Y}, \mathbf{Z}\) be a set of random variables. We say that \(\mathbf{X}\) is conditionally independent of \(\mathbf{Y}\) given \(\mathbf{Z}\) in a distribution \(P\) if \(P\) satisfies \(P(\mathbf{X} = x \perp \mathbf{Y}=y | \mathbf{Z}=z)\) for all values of \(x, y, z \in (Val(\mathbf{X}), Val(\mathbf{Y}), Val(\mathbf{Z}))\). If the set \(\mathbf{Z}\) is empty, we write \((\mathbf{X} \perp \mathbf{Y})\) and say that \(\mathbf{X}\) and \(\mathbf{Y}\) are marginally independent.
The distribution \(P\) satisfies \((\mathbf{X} \perp \mathbf{Y} | \mathbf{Z})\) IFF \(P(\mathbf{X}, \mathbf{Y} | \mathbf{Z}) = P(\mathbf{X}| \mathbf{Z}) P(\mathbf{Y} | \mathbf{Z})\)
- Symmetry: \[(\mathbf{X} \perp \mathbf{Y} | \mathbf{Z}) \implies (\mathbf{Y} \perp \mathbf{X} | \mathbf{Z})\]
- Decomposition: \[(\mathbf{X} \perp \mathbf{Y}, \mathbf{W} | \mathbf{Z}) \implies (\mathbf{X} \perp \mathbf{Y} | \mathbf{Z}), \;(\mathbf{X} \perp \mathbf{W} | \mathbf{Z})\]
- Weak Union: \[(\mathbf{X} \perp \mathbf{Y}, \mathbf{W} | \mathbf{Z}) \implies (\mathbf{X} \perp \mathbf{Y} | \mathbf{Z}, \mathbf{W}), \; (\mathbf{X} \perp \mathbf{W} | \mathbf{Z}, \mathbf{Y})\]
- Contraction: \[(\mathbf{X} \perp \mathbf{W} | \mathbf{Z}, \mathbf{Y}) \cap (\mathbf{X} \perp \mathbf{Y} | \mathbf{Z}) \implies (\mathbf{X} \perp \mathbf{Y}, \mathbf{W}| \mathbf{Z})\]
We can represent complicated probabilistic models using diagrammatic representations of probability distributions called probabilistic graphical models. These offer several useful properties:
- They provide a simple way to visualize the structure of a probabilistic model and can be used to design and motivate new models.
- Insights into the properties of the model, including conditional independence properties can be obtained by inspection of the graph.
- Complex computations, required to perform inference and learning in sophisticated models, can be expressed in terms of graphical manipulations, in which underlying mathematical expressions are carried along implicitly.
A probabilistic graphical model consists of:
- Nodes: each random variable (or group of random variables) is represented as a node in the graph
- Edges (links): links express probabilistic relationship between these random variables, the edges encode our intuition about the way the world works.
- Directed graphical models: in which the edges of the graphs have a particular directionality indicated by arrows (Bayesian networks). Directed graphs are useful for expressing causal relationships between random variables.
- Undirected graphical models: in which the edges of the graph do not carry arrows and have no directional significance (Markov random fields). Undirected graphs are better suited to expressing soft constraints between random variables.
The graph then captures the way in which the joint distribution over all of the random variables can be decomposed into a product of factors each depending only on a subset of the variables.
Bayesian Networks
The DAG of random variables can be viewed in two very different ways (Also strongly equivalent):
- As a data structure that provides the skeleton for representing a joint distribution compactly in a factorized way.
- As a compact representation for a set of conditional independence assumptions about a distribution.
We can view the graph as encoding a generative sampling process executed by nature, where the value for each variable is selected by nature using a distribution that depends only on its parents. In other words, each variable is a stochastic function of its parents.
In general, there are many weak influences that we might choose to model, but if we put in all of them, the network can become very complex, such networks are problematic from a representational perspective.
Bayesian Network Represents a Joint Distribution Compactly in a Factorized Way
Consider first an arbitrary joint distribution defined by \(P(\mathbf{Z})\) over random vector \(\mathbf{Z} = <A, B, C>\), by product rule, we have:
\[P(\mathbf{Z}) = P(C| A, B) P(A, B) = P(C | A, B) P(B | A) P(A)\]
We now represent the right-hand side in terms of a simgple graphical model as follows:
- First, we introduce a node for each of the random variables \(A, B, C\) and associate each node with the corresponding conditional distribution on the right-hand side.
- Then, for each conditional distribution we add directed links to the graph from the nodes to the variables on which the distribution is conditioned.

If there is a link going from a node \(A\) to a node \(B\), then we say that node \(A\) is parent of node \(B\) and \(B\) is the child of node \(A\) (change ordering of the decomposition will change the graph).
We can extend the idea to joint distribution of \(K\) random variables given by \(P(X_1, ...., X_K)\). By repeated application of the product rule of the probability, this joint distribution can be written as a product of conditional distributions:
\[P(X_1, ...., X_K) = P(X_K | X_{K-1}, ..., X_{1}) ... P(X_2 | X_1) P(X_1)\]
We can generate a graph similar to three-variable case, each node having incoming links from all lower numbered nodes. We say this graph is fully connected because there is a link between every pair of nodes. However, it is the absence (not fully connected) of links in the graph that conveys interesting information about the properties of the class of distributions that the graph represents.
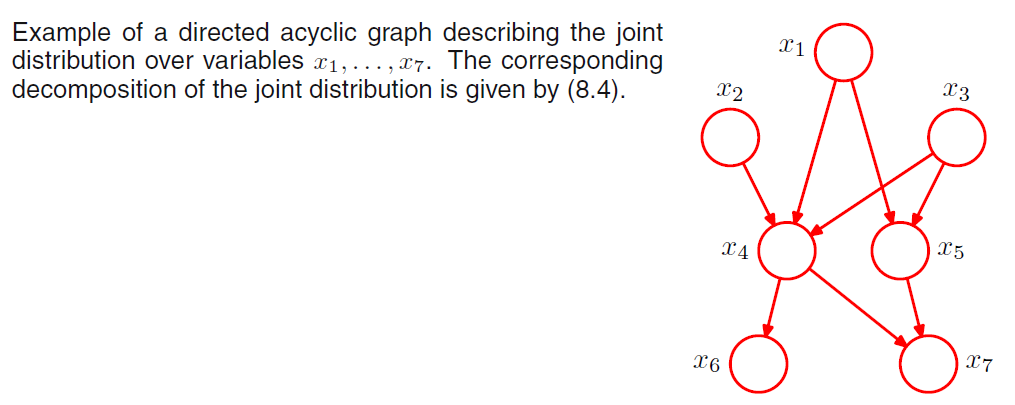
We can now state in general terms the relationship between a given directed graph and the corresponding distribution over the variables. Thus, for a graph with K nodes \(\mathbf{X} = <X_1, ...., X_K>\), the joint distribution is given by:
\[P(\mathbf{X}) = \prod^K_{k=1} P(X_k | \text{Parent}(X_k))\]
Where \(\text{Parent}(X_k)\) denotes the set of parents of \(X_k\).
Notice that, the directed graphs that we are considering are subject to an important restriction namely that there must be no directed cycles, that is, we are working with directed acyclic graphs or DAGs.
Example: Generative Models
There are many situations in which we wish to draw samples from a given probability distribution. One technique which is particularly relevant to graphical models is called ancestral sampling.
Consider a joint distribution \(P(\mathbf{X}), \mathbf{X} = <X_1, ...., X_K>\) that factorizes into a DAG. We shall suppose that the variables have been ordered from \(X_1\) to \(X_K\), in other words each node has a higher index than any of its parents. Our goal is to draw samples \(\hat{X}_1, ..., \hat{X}_K\) from the joint distribution.
To do this, we start from \(X_1\), and draw sample \(\hat{X}_1\) from the distribution \(P(X_1)\). We then work through each of the nodes in order, so that for node \(n\) we draw a sample from the conditional distribution \(P(X_n | \text{Parent}(X_n))\), in which the parent variables have been set to their sampled values.
To obtain a sample from some marginal distribution corresponding to a subset of the random variables, we simply take the sampled values for the required nodes and discard the rest. For example, to draw a sample from the distribution \(P(X_2, X_4)\), we simply sample from the full joint distribution and then retain the values \(\hat{X}_2, \hat{X}_4\) and discard the remaining values.
For practical applications of probabilistic models, it will typically be the higher-numbered variables corresponding to terminal nodes of the graph that represent the observations, with lower-numbered nodes corresponding to latent variables. The primary role of the latent variables is to allow a complicated distribution over the observed variables to tbe represented in terms of a model constructed from simpler conditional distributions.
Consider an object recognition task in which each observed data point corresponds to an image of on of the objects (vector of pixels). In this case, we can have latent variables be position and orientation of the object. Given a particular observed image, our goal is to find the posterior distribution over objects in which we integrate over all possible positions and orientations.  Given object, position, orientation, we can sample from the conditional distribution of image and generate pixels.
Given object, position, orientation, we can sample from the conditional distribution of image and generate pixels.
The graphical model captures causal process by which the observed data was generated. For this reason, such models are often called generative models.
Independence in Bayesian Networks
Our intuition tells us that the parents of a variable “shield” it from probabilistic influence that is causal in nature. In other words, once I know the value of the parents, no information relating directly or indirectly to its parents or other ancestors can influence my beliefs about it. However, information about its descendants can change my beliefs about it, via an evidential reasoning process.
Bayesian Network Semantics
Definition 3.1: Bayesian Network Structure
A Bayesian network structure \(G\) is a directed acyclic graph whose nodes represent random variables \(X_1, ...., X_n\). Let \(Pa^{G}_{X_i}\) denote the parents of \(X_i\) in \(G\), and \(\text{NonDescendants}_{X_i}\) denote the variables in the graph that are not descendants of \(X_i\). Then \(G\) encodes the following set of conditional independence assumptions, called local independence, and denoted by \(I_l (G)\):
\[I_l (G) = \{\text{For each variable $X_i$: ($X_i \perp \text{NonDescendants}_{X_i} | Pa^G_{X_i}$})\}\]
In other words, the local independence state that each node \(X_i\) is conditionally independent of its non-descendants given its parents.
Graphs and Distributions
We now show that the previous two definitions of BN are equivalent. That is, a distribution \(P\) satisfies the local independence associated with a graph \(G\) IFF \(P\) is representable as a set of CPDs associated with the graph \(G\).
I-Maps
Definition 3.2: Independencies in \(P\)
Let \(P\) be a distribution over \(\mathbf{X}\). We define \(I(P)\) to be the set of independence assertions of the form \((\mathbf{X} \perp \mathbf{Y} | \mathbf{Z})\)
Definition 3.3: I-Map
Let \(K\) be any graph object associated with a set of independencies \(I(K)\). We say that \(K\) is an I-map for a set of independencies \(I\) if \(I(K) \subseteq I\).
We now say that \(G\) is a I-map for \(P\) if \(G\) is an I-map for \(I(P)\). That is \(I(G) \subseteq I(P)\).
That is, for \(G\) to be an I-map of \(P\), it is necessary that \(G\) does not mis-lead us regarding independencies in \(P\): any dependence that \(G\) asserts must also hold in \(P\). Conversely, \(P\) may have additional independencies that are not reflected in \(G\).
I-Map to Factorization
A BN structure \(G\) encodes a set of conditional independence assumptions, every distribution for which \(G\) is an I-map must satisfy these assumptions.
Definition 3.4: Factorization (Chain Rule of Bayesian Network)
Let \(G\) be a BN graph over the variables \(X_1, ...., X_n\). We say that a distribution \(P\) over the same space factorizes according to \(G\) if \(P\) can be expressed as a product:
\[P(X_1, ...., X_n) = \prod^n_{i=1} P(X_i | Pa^G_{X_i})\]
Definition 3.5: Bayesian Network
A Bayesian network is a pair \(B = (G, P)\) where \(P\) factorizes over \(G\), and where \(P\) is specified as a set of CPDs associated with \(G\)'s nodes. The distribution \(P\) is often annotated \(P_B\).
Theorem 3.1: I-Map Factorization
Let \(G\) be a BN structure over a set of random variables \(\mathbb{X}\), and let \(P\) be a joint distribution over the same space. If \(G\) is a I-map for \(P\), then \(P\) factorizes according to \(G\) (can be written in the form as in definition 3.4).
Factorization to I-Map
Theorem 3.2: Factorization to I-Map
Let \(G\) be a BN structure over a set of random variables \(\mathbb{X}\) and let \(P\) be a joint distribution over the same space. If \(P\) factorizes according to \(G\), then \(G\) is an I-map for \(P\).
Independencies in Graphs
Knowing only that a distribution \(P\) factorizes over \(G\), we can conclude that it satisfies \(I_l (G)\). Are there other independencies that hold for every distribution \(P\) that factorizes over \(G\)? Our goal is to understand when we can guarantee that an independence \((\mathbf{X} \perp \mathbf{Y} | \mathbf{Z})\) holds in a distribution associated with a BN structure \(G\).
D-Separation
When influence can flow from \(X, Y\) via \(Z\) (\(X, Y\) are correlated), we say that the trail is active
- Direct connection: When \(X, Y\) are directly connected via edge. For any network structure \(G\) they are always correlated regardless of any evidence about any of the other variables in the network.
- Indirect connection: \(X, Y\) are not directly connected via edge, but there is a trail between then in the graph via \(Z\).
- Indirect causal effect: \(X \rightarrow Z \rightarrow Y\). If we observe \(Z\), then \(X, Y\) are conditionally independent, if \(Z\) is not observed, \(X\) influences \(Y\) by first sampling from \(P(X)\) then sampling \(Z\) from \(P(Z | X)\), so \(X, Y\) are not independent. (Active IFF \(Z\) is not observed)
- Indirect evidential effect: \(Y \rightarrow Z \rightarrow X\), same as indirect causal effect. (Active IFF \(Z\) is not observed)
- Common cause: \(X \leftarrow Z \rightarrow Y\), same as above, \(X\) can influence \(Y\) via \(Z\) IFF \(Z\) is not observed. (Active IFF \(Z\) is not observed)
- Common effect: \(X \rightarrow Z \leftarrow Y\), if \(Z\) is unobserved, then \(X, Y\) are independent, if it is observed then they are not. (Active IFF \(Z\) or one of \(Z\)'s descendants is observed)
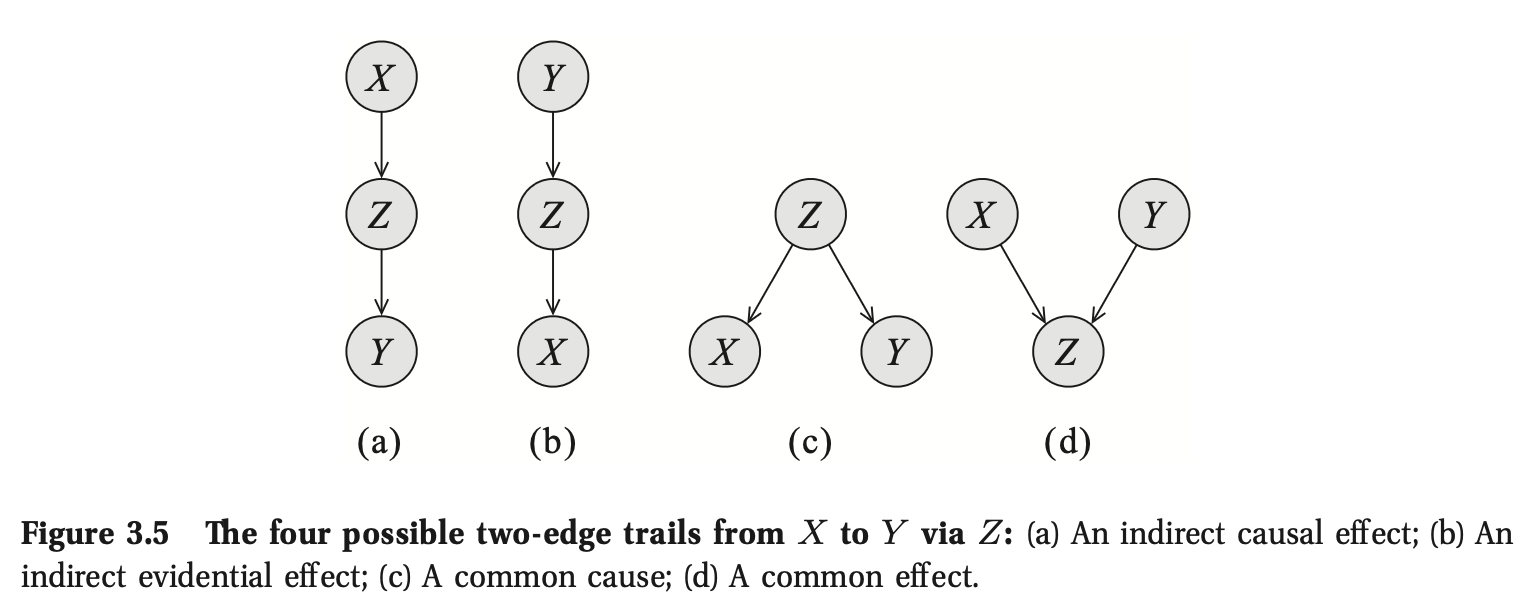
Definition 3.6: General Case (One path from \(X_1\) to \(X_n\))
Let \(G\) be a BN structure, and \(X_1 \rightleftharpoons .... \rightleftharpoons X_n\) a trail in \(G\). Let \(\mathbf{Z}\) be a subset of observed variables. The trail \(X_1 \rightleftharpoons .... \rightleftharpoons X_n\) is active given \(\mathbf{Z}\) if:
- Whenever we have a \(v\)-structure (Common effect), then \(X_i\) or one of its descendants are in \(\mathbf{Z}\).
- No other node along the trail is in \(\mathbf{Z}\).
Note if \(X_1\) or \(X_n\) are in \(\mathbf{Z}\), the trail is not active.
Definition 3.7: D-seperation
Let \(\mathbf{X}, \mathbf{Y}, \mathbf{Z}\) be three sets of nodes in \(G\). We say that \(\mathbf{X}, \mathbf{Y}\) are d-seperated given \(\mathbf{Z}\), denoted \(d-sep_G(\mathbf{X}; \mathbf{Y} | \mathbf{Z})\), if there is no active trail between any node \(X \in \mathbf{Z}\) and \(Y \in \mathbf{Y}\) given \(\mathbf{Z}\). We use \(I(G)\) to denote the set of independencies that correspond to d-separation:
\[I(G) = \{(\mathbf{X} \perp \mathbf{Y} | \mathbf{Z}) : d-sep_{G} (\mathbf{X}; \mathbf{Y} | \mathbf{Z})\}\]
The independencies in \(I(G)\) are precisely those that are guaranteed to hold for every distribution over \(G\).
Soundness and Completeness
Theorem 3.3: Soundness
If a distribution \(P\) factorizes according to \(G\), then \(I(G) \subseteq I(P)\).
In other words, any independence reported by d-separation is satisfied by the underlying distribution.
Definition 3.8: Faithful
A distribution \(P\) is faithful to \(G\) if, whenever \((X \perp Y | \mathbf{Z}) \in I(P)\), then \(d-sep_G(X; Y | \mathbf{Z})\).
In other words, any independence in \(P\) is reflected in the d-separation properties of the graph.
Theorem 3.4: Completeness
Let \(G\) be a BN structure. If \(X, Y\) are not d-separated given \(\mathbf{Z}\) in \(G\), then \(X, Y\) are dependent given \(\mathbf{Z}\) in some distribution \(P\) that factorizes over \(G\).
THese results state that for almost all parameterizations \(P\) of the graph \(G\), the d-separation test precisely characterizes the independencies that hold for \(P\).
Conditional Independence
An important concept for probability distributions over multiple variables is that of conditional independence. Consider three random variables \(A, B, C\) and suppose that the conditional distribution of \(A\), given \(B, C\) is such that it does not depend on the value of \(B\), so that:
\[P(A | B, C) = P(A | C)\]
Then:
\[P(A, B | C) = P(A | B, C) P(B | C) = P(A | C) P (B | C)\]
Thus, we can see that \(A, B\) are statistically independent given \(C, \; \forall C\). Note that this definition of conditional independence will require the above equation holds for all values fo \(C\) and not just for some values. The shorthand notation for conditional independence is:
\[A \perp \!\!\! \perp B \;|\; C\]
An important and elegant feature of graphical models is that conditional independence properties of the joint distribution can be read directly from the graph without having to perform any analytical manipulations. The general framework for achieving this is called d-seperation (d stands for directed).
Three example graphs
We start by illustrating the key concepts of d-separation by three motivating examples.
\(P(A, B, C) = P(A | C) P(B | C) P(C)\)  \(A, B\) are generally not statistically independent. However, we can easily see that \(A, B\) are conditionally independent given \(C\): \[P(A, B | C) = \frac{P(A, B, C)}{P(C)} = P(A | C) P (B | C)\]
\(A, B\) are generally not statistically independent. However, we can easily see that \(A, B\) are conditionally independent given \(C\): \[P(A, B | C) = \frac{P(A, B, C)}{P(C)} = P(A | C) P (B | C)\] 
We can provide a simple graphical interpretation of this result by considering the path from node \(A\) to node \(B\) via \(C\). The node \(C\) is said to be tail-to-tail with respect to this path because the node is connected to the tails of the two arrows. However, when we condition on node \(C\) (observed \(C\)), the conditional node blocks the path from \(A\) to \(B\) so causes then to become conditionally independent.
\(P(A, B, C) = P(B | C) P(C | A) P (A)\)  \(A, B\) are generally not statistically independent. However, we can easily see that \(A, B\) are conditionally independent given \(C\) by: \[P(A, B | C) = \frac{P(A, B, C)}{P(C)} = P(A | C) P (B | C)\]
\(A, B\) are generally not statistically independent. However, we can easily see that \(A, B\) are conditionally independent given \(C\) by: \[P(A, B | C) = \frac{P(A, B, C)}{P(C)} = P(A | C) P (B | C)\]
We can provide a simple graphical interpretation of this result by considering the path from node \(A\) to node \(B\) via \(C\). The node \(C\) is said to be head-to-tail with respect to this path because the node is connected to the head and tail of the two arrows. However, when we condition on node \(C\) (observed \(C\)), the conditional node blocks the path from \(A\) to \(B\) so causes then to become conditionally independent.
\(P(A, B, C) = P(A)P(B)P(C | A, B)\)  We can easily see that \(A, B\) are not conditionally independent. However, we can see that \(A, B\) are statistically independent: \(P(A, B) = \sum_{C} P(A)P(B)P(C | A, B) = P(A)P(B)\)
We can easily see that \(A, B\) are not conditionally independent. However, we can see that \(A, B\) are statistically independent: \(P(A, B) = \sum_{C} P(A)P(B)P(C | A, B) = P(A)P(B)\)
Thus, our third example has the opposite behaviour from the first two. The node \(C\) is said to be head-to-head with respect to this path because the node is connected to the heads of the two arrows. When the node \(C\) is not given (unobserved), it blocks the path so \(A\) and \(B\) is independent, however, when the node \(C\) is given, \(A\) and \(B\) becomes dependent.
There is one more relationship associate with third example. First we say that node \(Y\) is a descendant of node \(X\) if there is a path from \(X\) to \(Y\) in which each step of the path follows the directions of the arrows. Then it can be shown that a head to head path will become unblocked if either the node or any of its descendants is observed.
D-separation
Consider a general directed graph in which \(A, B, C\) are arbitrary sets of nodes. We wish to ascertain whether a particular conditional independence statement \(A \perp \!\!\! \perp B \;|\; C\) is implied by a given directed acyclic graph. To do so, we consider all possible paths from any node in \(A\) to any node in \(B\). Any such path is said to be blocked if it includes a node such that either:
- The arrows on the path meet either head-to-tail or tail-to-tail at the node, and the node is in the set \(C\).
- The arrows meet head-to-head at the node, and neither the node, nor any of its descendants, is in the set \(C\).
If all paths are blocked, then \(A\) is said to be d-separated from \(B\) by \(C\), and the joint distribution over all of the variables in the graph will satisfy \(A \perp \!\!\! \perp B \;|\; C\).
Consider the problem of finding the posterior distribution for the mean of an univariate Gaussian distribution. This can be represented by the directed graph in which the joint distribution is defined by a prior \(P(\mu)\) and \(P(\mathbf{X} | \mu)\) to form the posterior distribution: \[P(\mu | \mathbf{X}) = P(\mu) P(\mathbf{X} | \mu) \] 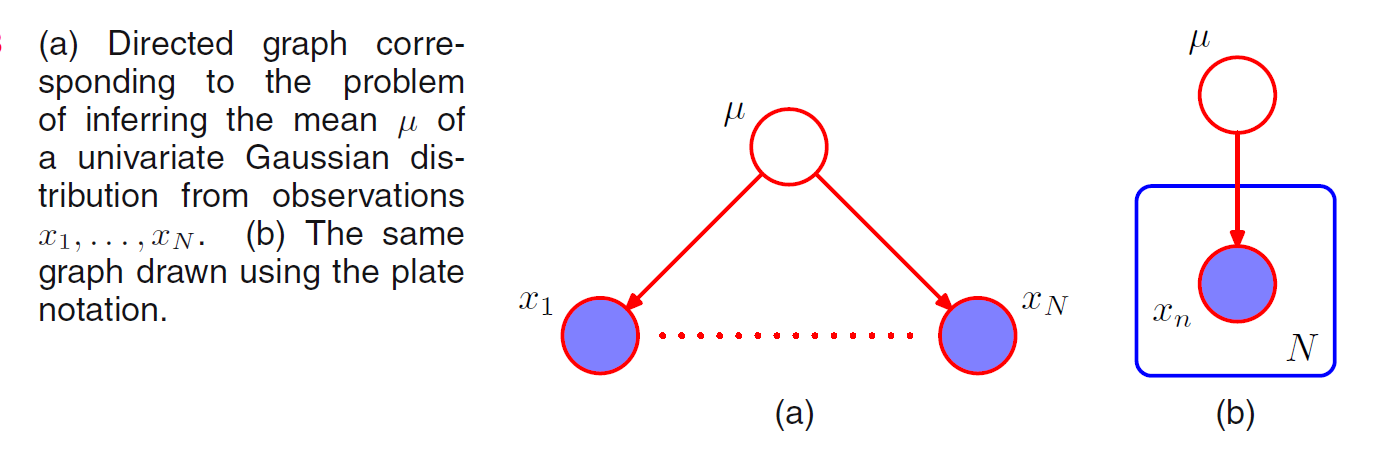 In practice, we observe \(D = \{X_1, ...., X_N\}\) with conditional distribution \(P(X_1 | \mu) , ...., P(X_N | \mu)\) respectively, and our goal is to infer \(\mu\). Using d-separation, we note that there is a unique path from any \(X_i\) to any other \(X_{j\neq i}\) and that this path is tail-to-tail with respect to the observed node \(\mu\). Every such path is blocked and so the observations \(D=\{X_1, ..., X_N\}\) are independent given \(\mu\): \[P(\mathbf{X} | \mu) = \prod^N_{i=1} P(X_i | \mu)\] However, if we do not conditional on \(\mu\), the data samples are not independent: \[P(\mathbf{X}) = \int_{\mu} P(\mathbf{X} | \mu) P(\mu) \neq \prod^N_{i=1} P(X_i)\]
In practice, we observe \(D = \{X_1, ...., X_N\}\) with conditional distribution \(P(X_1 | \mu) , ...., P(X_N | \mu)\) respectively, and our goal is to infer \(\mu\). Using d-separation, we note that there is a unique path from any \(X_i\) to any other \(X_{j\neq i}\) and that this path is tail-to-tail with respect to the observed node \(\mu\). Every such path is blocked and so the observations \(D=\{X_1, ..., X_N\}\) are independent given \(\mu\): \[P(\mathbf{X} | \mu) = \prod^N_{i=1} P(X_i | \mu)\] However, if we do not conditional on \(\mu\), the data samples are not independent: \[P(\mathbf{X}) = \int_{\mu} P(\mathbf{X} | \mu) P(\mu) \neq \prod^N_{i=1} P(X_i)\]
Markov Random Fields
Directed Graphical models specify a factorization of the joint distribution over a set of variables into a product of local conditional distributions. They also defined a set of conditional independence properties that must be satisfied by any distribution that factorizes according to the graph. A Markove random field has:
- A set of nodes each of which corresponds to a random variable or group of random variables
- A set of links each of which connects a pair of nodes. The links are undirected that is they do not carry arrows.
Conditional Independence Properties
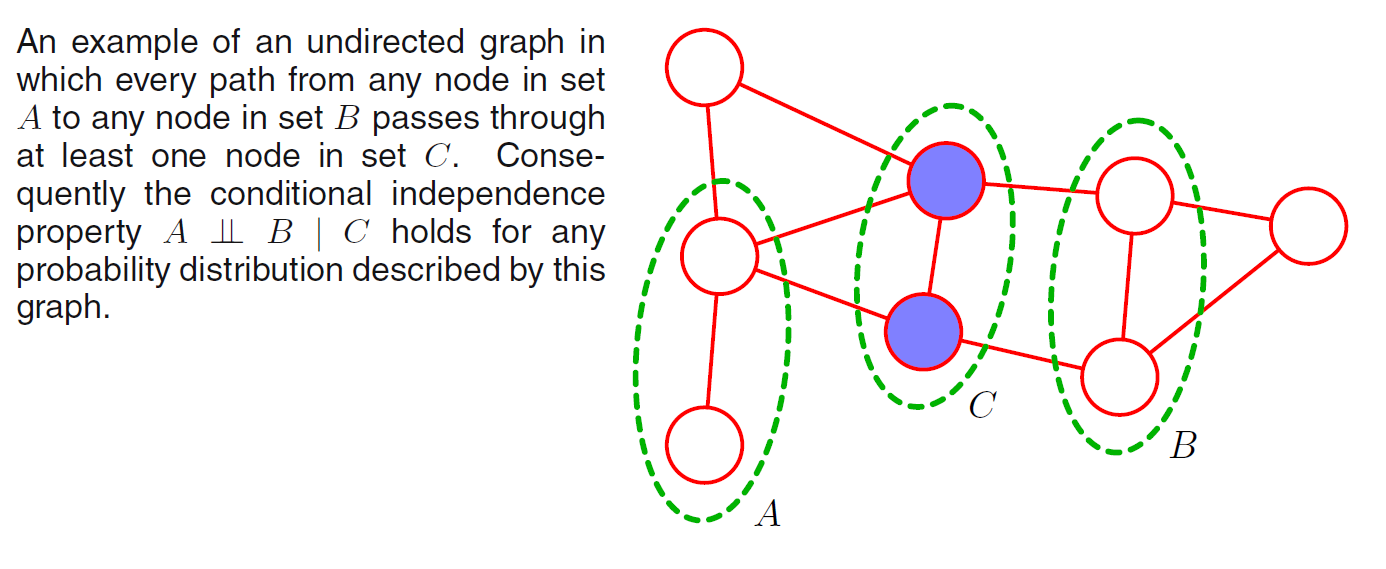
Testing for conditional independence in undirected graph is simpler than in directed graph. Let \(A, B, C\) be three sets of nodes and we consider the conditional independence property \(A \perp \!\!\! \perp B \;|\; C\). To test whether this property is satisfied by a probability distribution defined by the graph:
- Consider all possible paths that connect nodes in set \(A\) to nodes in set \(B\). If all such paths pass through one or more nodes in set \(C\), then all such paths are blocked and so the conditional independence properties holds. If there is at least one such path that is not blocked, then there will exist at least some distributions corresponding to the graph that do not satisfy this conditional independence relation.
Factorization Properties
We now express the joint distribution \(P(\mathbf{X})\) as a product of functions defined over set of random variables that are local to the graph.
If we consider two nodes \(X_i\) and \(X_j\) that are not connected by a link, then these variables must be conditionally independent given all other nodes in the graph, because there is no direct path between the two nodes and all other paths are blocked:
\[P(X_i, X_j | \mathbf{X}_{k\notin \{i, j\}}) = P(X_i | \mathbf{X}_{k\notin \{i, j\}}) P(X_j | \mathbf{X}_{k\notin \{i, j\}})\]
A clique is a subset of nodes in a graph such that there exists a link between all pairs of nodes in the subset. In other words, the nodes in the set are fully connected. Furthermore, a maximal clique is a clique such that it is not possible to include any other nodes from the graph in the set without it ceasing to be a clique.

We can therefore define the factors in the decomposition of the joint distribution to be functions of the variables in the cliques. In fact, we can consider functions of the maximal cliques, without loss of generality because other cliques must be subsets of maximal cliques.
Let \(C\) be a clique and the set of random variables in that clique by \(\mathbf{X}_C\). Then the joint distribution is written as a product of potential functions \(\psi_C(\mathbf{x}_C) \geq 0\) over the maximal cliques of the graph:
\[P(\mathbf{X}) = \frac{1}{Z} \prod_{C} \psi_{C} (\mathbf{X}_C)\]
Here the quantity \(Z\) is called partition function which is used for normalization to ensure the result is a proper joint distribution:
\[Z = \sum_{X} \prod_{C} \psi_{C} (\mathbf{X}_C)\]
In directed graph, we have the links to be conditional distribution, in undirected graph, we do not restrict the choice of potential functions.
Ref
PRML chapter 8
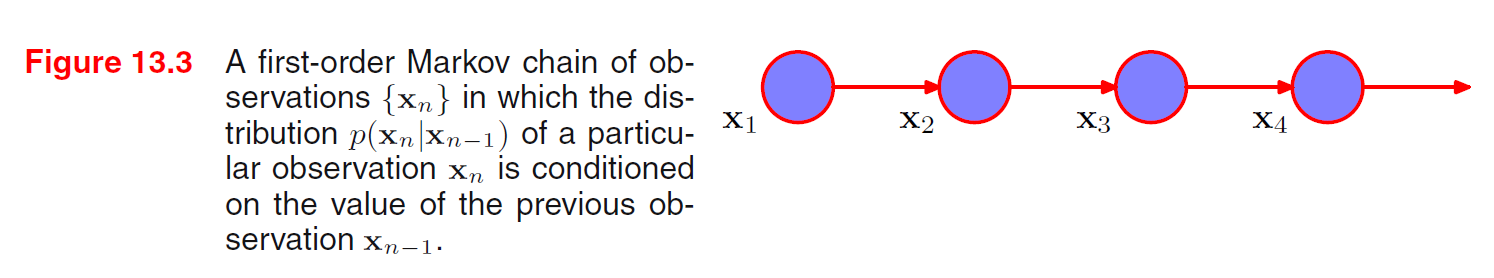
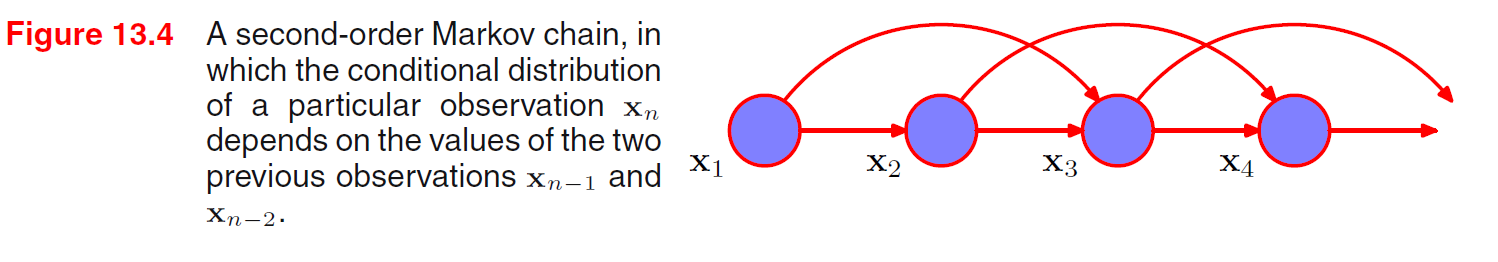
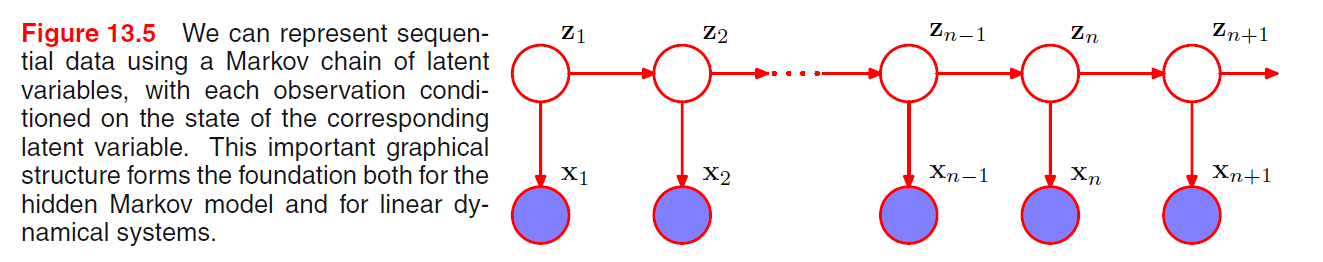


 Given object, position, orientation, we can sample from the conditional distribution of image and generate pixels.
Given object, position, orientation, we can sample from the conditional distribution of image and generate pixels.



 We can easily see that
We can easily see that  In practice, we observe
In practice, we observe